1. Background
Nearly one-third of the global population suffers from anemia (1), with iron deficiency anemia (IDA) representing 50% of all cases, making it the most prevalent type of anemia (2). Iron deficiency anemia affects over 1.2 billion people worldwide (3) and is associated with adverse pregnancy outcomes, impaired cognitive development in children and the elderly (4, 5), and decreased physical capacity and work productivity in adults, which in turn poses significant challenges to societal and economic development (6). Common causes of IDA include increased iron demand, chronic blood loss, insufficient iron intake, and impaired iron absorption, with inadequate iron intake being the primary contributor (7).
Numerous studies have highlighted the impact of various factors on iron absorption, particularly the potential role of Helicobacter pylori infection in the development of IDA, a topic that remains a subject of ongoing debate. Helicobacter pylori is a Gram-negative bacterium that colonizes the human gastric epithelium (4) and is one of the most common chronic infectious pathogens in humans (8). Its genome, comprising approximately 1,500 genes, exhibits significant variability, allowing the bacterium to adapt to the complex gastric environment and establish long-term colonization in the mucosa (9). Key virulence factors of H. pylori, including cytotoxin-associated gene A, vacuolating cytotoxin A, and urease, contribute to gastric pathology by disrupting host cell functions and compromising the gastric mucosal barrier (10). Over half of the global population is infected with H. pylori (11).
Observational studies conducted between 2003 and 2010 have shown a correlation between H. pylori infection and the occurrence of IDA (12-14). Case reports from 2007 to 2014 further suggested that eradicating H. pylori could improve hemoglobin (HGB), iron, and ferritin levels, thereby treating IDA (15, 16). However, some studies argue that H. pylori infection only increases the risk of IDA in specific patient groups, such as those with gastrointestinal ulcers or bleeding (17, 18).
Retrospective studies and cohort studies conducted between 2017 and 2019 demonstrated no significant association between H. pylori infection alone and IDA, implying that H. pylori eradication may be unnecessary for IDA patients without gastrointestinal ulcers or bleeding (19-22). Nevertheless, in clinical practice, it remains challenging to completely rule out the presence of ulcers or bleeding in IDA patients. Therefore, scientifically rigorous study designs are required to further investigate whether H. pylori infection alone contributes to the development of IDA.
Mendelian randomization (MR) is an epidemiological method that strengthens causal inference (23). By utilizing single-nucleotide polymorphisms (SNPs) as instrumental variables (IVs), MR leverages genetic variants that are randomly distributed and unaffected by environmental factors or confounders (24), making it a robust tool for investigating causal relationships.
2. Objectives
In this study, we performed a two-sample MR analysis to investigate the causal effect of H. pylori infection on the risk of IDA.
3. Methods
3.1. Research Design
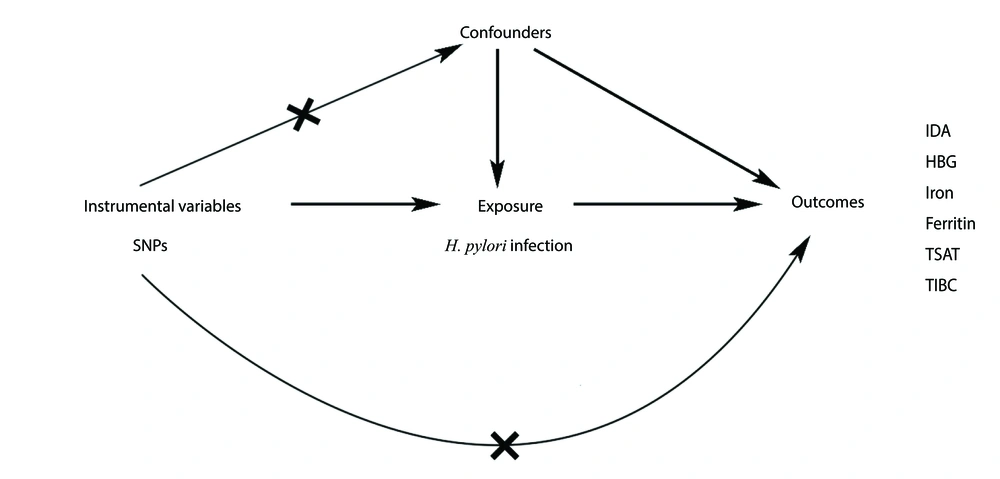
Two-sample MR methods were used in this study, with genetic data derived from genome-wide association studies (GWAS) that followed the defined guidelines for MR analysis (25). The research design was based on the following primary assumptions: (1) Genetic variation was significantly associated with H. pylori infection; (2) genetic variation was not associated with any of the potential confounding factors; and (3) genetic variation was not associated with IDA unless caused by H. pylori infection (Figure 1).
Schematic representation of Mendelian randomization (MR) study on the relationship between Helicobacter pylori infection and outcomes. This MR study was based on the hypothesis that the instrumental variable (IV) was correlated to H. pylori infection but not with any confounding factors, and that the IV only influenced the risk of IDA and related serum indicators through H. pylori infection. Abbreviations: IDA, iron deficiency anemia; HBG, hemoglobin; TSAT, transferrin saturation; TIBC, total iron-binding capacity.
3.2. Instrumental Variable Selection
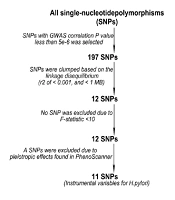
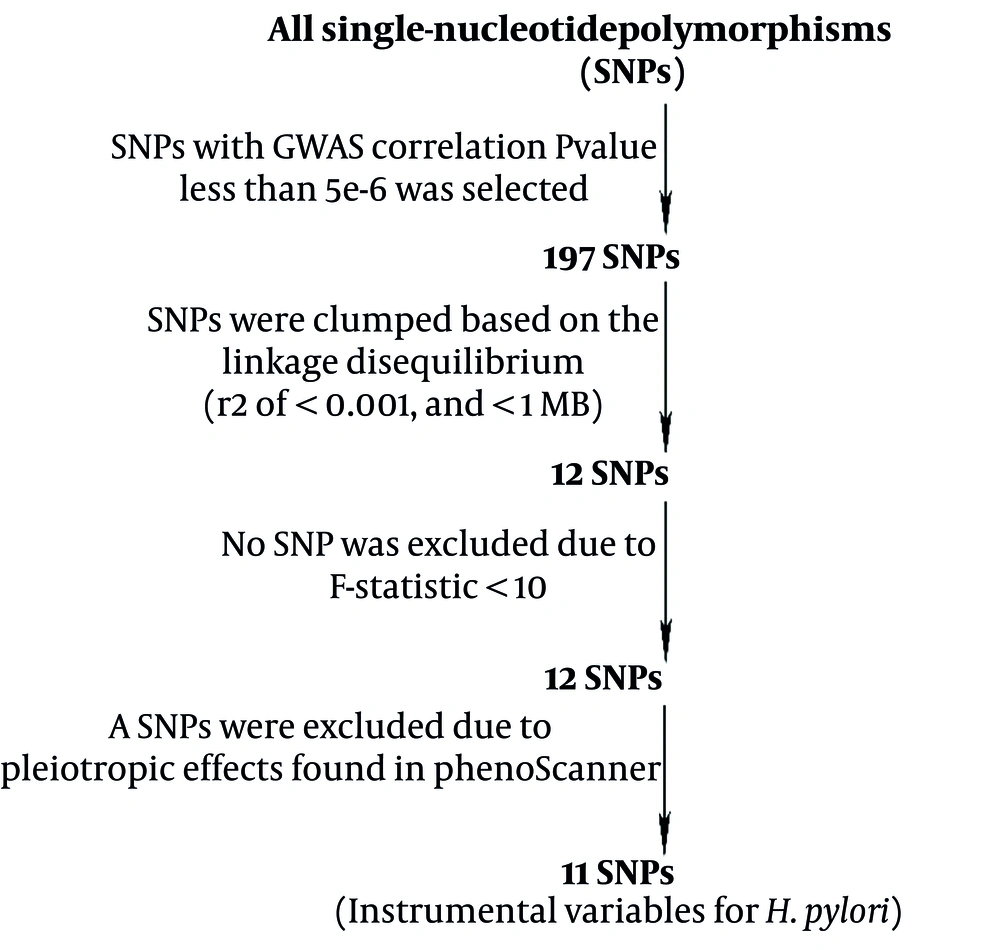
To determine if the genetic IVs that satisfied the MR hypothesis criteria were eligible for this study, the GWAS summary data on H. pylori infections obtained from the European Bioinformatics Institute (EBI) database were subjected to a series of quality control measures. The GWAS-related P-value of 5e-8 was insufficient to screen a sufficient number of feature-related SNPs; hence, a lenient standard of 5e-6 was applied (26). Thereafter, SNPs with high linkage disequilibrium (LD) were filtered using the LD clustering algorithm (
3.3. Outcome Data Sources
The GWAS Catalog database provided download access to the IDA dataset (GCST90038659), which included 2,941 cases and 481,657 controls (29), in addition to the serum HGB concentration dataset (GCST90002384), which included 408,112 participants of European ancestry (30). Furthermore, GWAS datasets related to serum iron metabolism-related indices were obtained from a meta-analysis of three GWAS in Iceland, the United Kingdom, and Denmark, which included serum iron (N = 135,430), ferritin (N = 246,139), transferrin saturation (TSAT) (N = 131,471), and total iron-binding capacity (TIBC) (N = 135,430) (31). Detailed information about all datasets, as well as instructions for obtaining them, are provided in Appendix 1 in Supplementary File.
3.4. Mendelian Randomization Analysis
In this study, four different MR approaches were used, including inverse-variance weighted (IVW), weighted median, MR-Egger, and MR-Pleiotropy Residual Sum and Outlier (MR-PRESSO) methods. The IVW estimates served as the primary outcome; however, MR-Egger, weighted median, and MR-PRESSO methods were used to improve the IVW estimates because, despite their lower efficiency, they may be more reliable in a wider range of scenarios. Furthermore, funnel plots were used to investigate the potential directional pleiotropic effects, whereas the MR-Egger intercept test was used to assess horizontal pleiotropic effects, the Cochran’s Q test was used to identify heterogeneity, and the MR-PRESSO method was used to identify and correct horizontal pleiotropic effects to confirm IVW results. Additionally, a leave-one-out analysis was performed to assess the impact of individual SNP bias on the MR results.
3.5. Statistical Analysis
All data were analyzed using the Two Sample MR package (ver. 0.5.6) in R software (ver. 4.2.0). A Bonferroni-corrected P-value of 0.0083 (0.05/6) was considered statistically significant.
4. Results
4.1. Screening of Instrumental Variables
Based on GWAS data in the EBI database, 197 feature-related SNPs were screened, and after removing SNPs with high LD, 12 SNPs were identified with F-values of > 10. Furthermore, PhenoScanner was used to scan for the relevant characteristics of these SNPs. rs35030589 was found to be associated with IDA risk factors such as malabsorption, celiac disease, ulcers, rheumatoid arthritis, and BMI; thus, it was eliminated. Then, the remaining 11 SNPs were used as IVs for MR analysis (Figure 2 and Appendix 2 in Supplementary File).
4.2. Mendelian Randomization Analysis
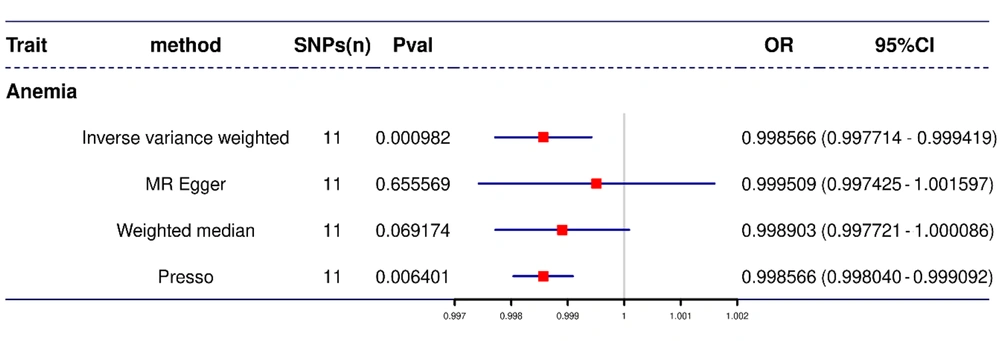
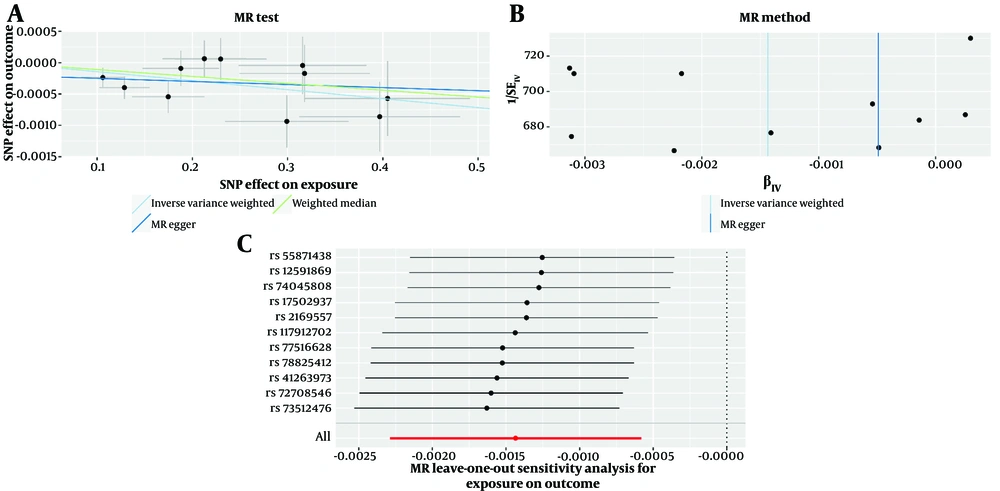
Mendelian randomization analysis using the IVW method revealed that H. pylori infection significantly reduced the risk of developing IDA by 0.14% [odds ratio (OR) = 0.9986, 95% confidence interval (CI) = 0.9977 - 0.9994, P = 98e-5]. This result was also supported by those obtained using the MR-PRESSO (OR = 0.9986, 95% CI = 0.9980 - 0.9991, P = 0.0064), weighted median (OR = 0.9989, 95% CI = 0.9977 - 1.0001, P = 0.07), and MR-Egger regression (OR = 0.9995, 95% CI = 0.9974 - 1.0016, P = 0.66) methods, which yielded similar risk estimates (Figure 3). Furthermore, the MR-Egger regression’s intercept yielded a P-value of 0.3566 (P > 0.05), indicating that there was no pleiotropy and thus proving that hypotheses 2 and 3 were correct. The MR-PRESSO heterogeneity study revealed no outliers (the global heterogeneity test yielded a P-value of 0.55), and Cochran’s Q test yielded a P-value of > 0.05 (P = 0.51), indicating no heterogeneity. Furthermore, the leave-one-out analysis revealed that these values were unaffected by individual SNP bias (Figure 4).
Mendelian randomization (MR)-Egger estimates of significant results from Helicobacter pylori on iron deficiency anemia (IDA), A, scatter plots from genetically predicted H. pylori on IDA; B, funnel plot from genetically predicted H. pylori on IDA; C, leave-one-out plot from genetically predicted H. pylori on IDA
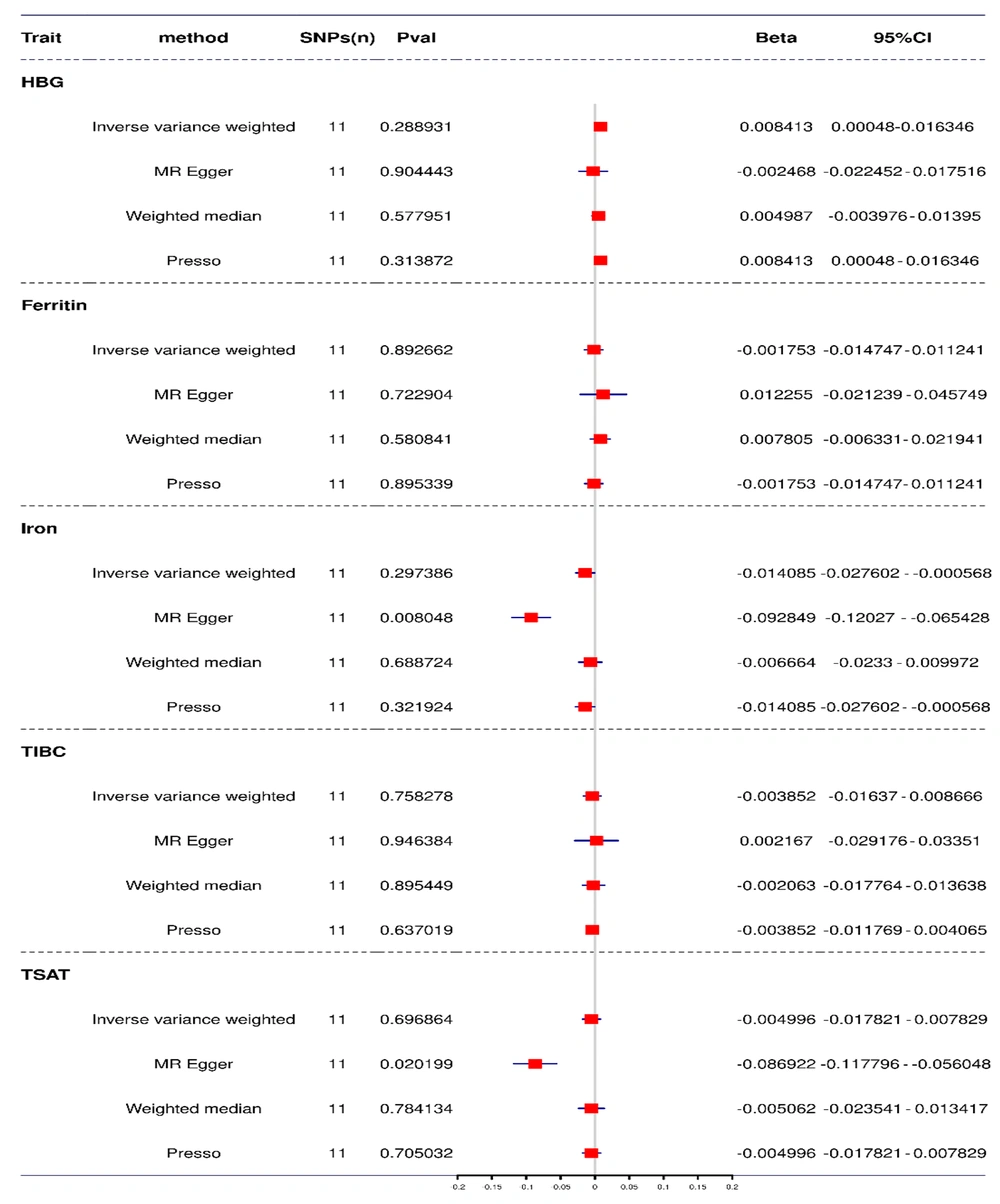
The MR analysis using the IVW method revealed no causal relationship between H. pylori infection and serum iron metabolism-related indices (HGB: Beta = 0.0084, 95% CI = 0.0005 - 0.0163, P = 0.29; iron: Beta = -0.0149, 95% CI = -0.0276 to -0.0006, P = 0.30; ferritin: Beta = -0.0018, 95% CI = -0.0147 - 0.0112, P = 0.90; TIBC: Beta = -0.0039, 95% CI = -0.0164 - 0.009, P = 0.76; TSAT: Beta = -0.0050, 95% CI = -0.0178 - 0.0078, P = 0.70) (Figure 5).
The results obtained using the other three secondary MR methods revealed that only the results of the MR-Egger test regarding the causal relationship between H. pylori infection and serum iron metabolism-related indices met the Bonferroni-corrected P-value standard (beta = -0.0928, 95% CI = -0.1203 - 0.0654, P = 0.0080) (Figure 5). However, the leave-one-out analysis revealed a significant bias in the results (shown in Appendices 3 - 5 in Supplementary File).
5. Discussion
Traditional research methods face significant limitations when exploring the relationship between H. pylori infection and IDA. Although some studies suggest that H. pylori infection is an independent risk factor for IDA (32), these studies failed to effectively control for confounding factors such as peptic ulcers and other gastrointestinal diseases. When these confounders were accounted for, the impact of H. pylori infection on IDA became insignificant. For example, two retrospective studies found that H. pylori infection did not increase the risk of unexplained IDA after excluding confounding factors such as peptic ulcers and autoimmune gastritis (19, 21). Additionally, while a household-randomized controlled trial in 2006 indicated that treating H. pylori infection could promote recovery from refractory IDA (33), a randomized controlled trial in 2008 showed no significant association between H. pylori eradication and IDA improvement (34). Further supporting this view, Saler et al. found that H. pylori infection was not associated with IDA in men with normal gastrointestinal endoscopic findings, suggesting that H. pylori may not play a critical role in the development of IDA in the absence of severe gastrointestinal pathologies (35).
Overall, traditional research methods struggle to fully eliminate the interference of confounding factors such as peptic ulcers and gastrointestinal bleeding, which may explain the inconsistencies in research findings. The discrepancy between the findings of this study and those of traditional research can primarily be attributed to the advantages of the MR approach. By using genetic variants as IVs, MR minimizes the influence of confounding factors on the relationship between exposure and outcome (36, 37).
In this study, we employed MR to investigate the causal relationship between H. pylori infection and IDA, resulting in a conclusion that significantly diverges from previous findings: Helicobacter pylori infection is not a causative factor for IDA and may, in fact, reduce the incidence of IDA. This conclusion was supported by multiple MR analytical methods, including IVW and MR-PRESSO analyses, both of which demonstrated an OR of less than 1 (OR = 0.9986) and achieved stringent statistical significance (P < 0.0083). Although the weighted median and MR-Egger methods did not reach statistical significance, they provided similar risk estimates. Furthermore, no abnormalities were detected in heterogeneity or pleiotropy tests, further reinforcing the reliability of the study's results.
The strengths of this study are highlighted by several key aspects. First, we are the first to apply MR to investigate the causal relationship between H. pylori infection and IDA, effectively addressing potential confounding factors and reverse causality often present in traditional observational studies. Second, we utilized summary-level data from multiple large genetic consortia, ensuring both the reliability of data sources and the adequacy of the sample size. Third, our analysis was focused on individuals of European ancestry, which minimizes the potential for population stratification bias, though it may limit the generalizability of the findings. Therefore, further validation in other populations is needed in future studies.
The potential mechanisms by which H. pylori infection may reduce the risk of IDA remain unclear, but several factors could be involved. First, H. pylori infection may indirectly affect iron absorption and metabolism by modulating the host's immune response or altering the gut microbiota (38). Second, H. pylori infection can increase gastric acid secretion (39, 40), lowering gastric pH and thereby enhancing the solubility and absorption of iron in the stomach (41). Additionally, H. pylori infection may directly or indirectly promote iron absorption or utilization through mechanisms that are yet to be fully understood. Further research is needed to validate these potential mechanisms and provide a clearer understanding of the complex relationship between H. pylori infection and IDA.
Our study provides genetic evidence that H. pylori infection does not increase the risk of IDA, prompting a more cautious approach to testing and treating H. pylori infection in patients with unexplained IDA. Beyond reducing unnecessary diagnostic costs, this approach also avoids potentially disrupting the unknown beneficial effects of H. pylori infection (42). For instance, evidence suggests that infection with certain H. pylori strains is associated with a reduced prevalence of Barrett's esophagus, esophageal adenocarcinoma (43), asthma, and atopic diseases (44). These findings indicate that H. pylori infection may confer protective effects under certain conditions. Therefore, clinical practice should carefully weigh its potential benefits against risks.
Although the findings of this study hold significant clinical implications, further research is needed to validate these results. Future studies should focus on elucidating the specific mechanisms by which H. pylori infection influences the development of IDA and should verify these findings through larger-scale MR analyses or randomized controlled trials. If these conclusions are further substantiated, they could have a profound impact on the management of H. pylori infection and IDA prevention strategies, particularly in populations without severe gastrointestinal pathologies. Additionally, future research should explore the effects of different H. pylori strains on IDA and investigate the potential protective roles of H. pylori infection across diverse populations, thereby providing more comprehensive guidance for clinical practice.
5.1. Conclusions
Helicobacter pylori infection is not a causative factor for IDA and may even act as a protective factor.