1. Background
Phospholipids are the fundamental building blocks of biological membranes, playing an essential role in maintaining cellular integrity and supporting various membrane-related functions (1, 2). As artificially constructed bilayer lipid vesicles, liposomes offer significant advantages for the controlled release of both hydrophobic and hydrophilic drugs, leading to improved drug bioavailability, enhanced targeting potential, and minimized side effects (3, 4). Liposomes, alongside other nanostructure-based systems, have significantly advanced the improvement of therapeutic outcomes in clinical applications for diverse pathological conditions (5, 6).
Despite their benefits, liposomes face several challenges, particularly in maintaining stability over time and ensuring that the targeting functional groups attached to phospholipid headgroups do not detach and thereby lose efficacy (2). This detachment process can severely impact the stability and targeting capabilities of liposomal formulations, highlighting the necessity for a deeper understanding of the molecular interactions within the bilayer (7). The presence of cholesterol and the purity of phospholipids significantly influence the physicochemical properties of liposomes, directly affecting their performance in drug delivery applications (8).
Cholesterol is frequently incorporated into liposomes to enhance membrane stability and reduce permeability (9). Mixed lipid systems can also modulate liposome properties by combining different phospholipids, thereby optimizing encapsulation efficiency, release kinetics, and cellular targeting (10). In particular, the inclusion of phosphatidylserine (PS) in the liposom formulations enhances recognition by specific cell receptors, facilitating more effective targeted drug delivery (11).
Molecular dynamic (MD) simulation has become an essential approach for studying lipid behavior in liposomal formulations at the atomistic level (12). By analyzing free energy profiles, researchers gain insights into how lipid composition and environmental factors influence membrane integrity under varying physiological conditions (13, 14). In this study, we use MD simulations with the umbrella sampling method to explore the effects of lipid purity and cholesterol incorporation on liposome stability. Bilayer models with varying lipid compositions were constructed, including pure 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE), mixtures of DSPE with cholesterol, as well as combinations of DSPE, 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), and 1,2-distearoyl-sn-glycero-3-phospho-L-serine (DSPS) both with and without cholesterol.
2. Objectives
The results from this study provide valuable insights for optimizing liposomal formulations to enhance stability and performance in targeted drug delivery applications.
3. Methods
3.1. Lipid Bilayer Construction
Atomistic models of bilayer membranes were built using the CHARMM-GUI web server (http://www.charmm-gui.org). The bilayer dimensions were set to 5 × 5 nm in the x-y plane and 15 nm in the z-direction to prevent steric interactions and ensure lipid detachment could occur without violating periodic boundary conditions. The system was solvated using the TIP3P water model, and sodium and chloride ions were added to neutralize the system.
3.2. Force Field and Parameters
All MD simulations were conducted using GROMACS 2021 with the CHARMM36 force field. Energy minimization was performed using the steepest descent algorithm (15). Periodic boundary conditions were applied in all directions, and long-range electrostatic interactions were computed using the Particle Mesh Ewald method (16). Van der Waals and short-range Coulombic interactions were truncated at a cutoff of 1.2 nm, and hydrogen bonds were constrained using the LINCS algorithm (17). The system was equilibrated in the NVT ensemble for 1 ns with a V-rescale thermostat maintaining the temperature at 310 K, followed by 5 ns of equilibration in the NPT ensemble using a Parrinello-Rahman barostat (18) to maintain the pressure at 1 bar.
3.3. Umbrella Sampling and Free Energy Calculations
An umbrella sampling technique was used to extract a single lipid molecule from the bilayer along the z-axis (19). Steered MD was used to create the initial configurations, pulling the lipid at a steady rate of 0.01 nm/ps. The 50 windows of the reaction coordinate, separated by 0.1 nm, covered the range from the equilibrated bilayer position to a fully detached state. Each window was subjected to a harmonic restraint with a force constant of 1000 kJ/mol/nm2. Simulations for each window were run for 20 ns. The weighted histogram analysis method (WHAM) was used to combine data from all windows and compute the potential of mean force (PMF). The following lipid bilayer compositions were investigated: S1: A hundred percent DSPE, S2: Seventy percent DSPE + 30% Cholesterol, S3: Fifty percent DSPE + 50% DSPC, S4: Thirty-five percent DSPE + 35% DSPC + 30% Cholesterol, S5: Fifty percent DSPE + 50% DSPS, S6: Thirty-five percent DSPE + 35% DSPS + 30% Cholesterol.
4. Results and Discussion
The primary objective of this study was to evaluate how variations in phospholipid composition and cholesterol incorporation influence the stability of phospholipids in liposomal bilayers. To this end, we analyzed six systems that differ in lipid purity and cholesterol content, considering DSPE as the main phospholipid, as it is the common anchored lipid with the targeting agents. The values of free energy (E), as well as the area under the curve of PMF and force diagrams, are summarized in Table 1.
| Phospholipid Composition | AUC of PMFs | AUC of Forces | Binding Energy (kcal/mol) |
|---|---|---|---|
| DSPE 100 (S1) | 126 | 79353 | 41.86 |
| DSPE 70 + Col 30 (S2) | 93 | 69039 | 34.47 |
| DSPE 50 + DSPC 50 (S3) | 89 | 59369 | 31.66 |
| DSPE 35 + DSPC 35 + Col 30 (S4) | 57 | 46555 | 28.83 |
| DSPE 50 + DSPS 50 (S5) | 115 | 75022 | 38.19 |
| DSPE 35 + DSPS 35 + Col 30 (S6) | 88 | 59054 | 31.43 |
Detachment Parameters of Phospholipids in Liposomal Systems
4.1. Binding Energy (E)
In the pure DSPE system (S1), the binding energy of lipids in the membrane is 41.86 kcal/mol, which signifies strong intermolecular interactions resulting from the tight packing of saturated lipid tails and the coulombic interactions between headgroups. When 30% cholesterol is incorporated (S2), the binding energy decreases significantly to 34.47 kcal/mol, a reduction of 17.63%. This decrease highlights cholesterol’s disruptive effect on the uniformity of lipid packing, reducing the overall cohesion between phospholipids. Similar trends are observed in the mixed lipid systems. The DSPE/DSPC mixture (S3) exhibits a binding energy of 31.66 kcal/mol, which further decreases to 28.83 kcal/mol in S4, a reduction of 8.94%. The DSPE/DSPS system shows a binding energy of 38.19 kcal/mol in S5, compared to 31.43 kcal/mol in S6, a reduction of 17.69% after cholesterol addition in this system. The consistent reduction in binding energy with cholesterol incorporation suggests a general pattern whereby cholesterol facilitates lipid detachment from the membrane.
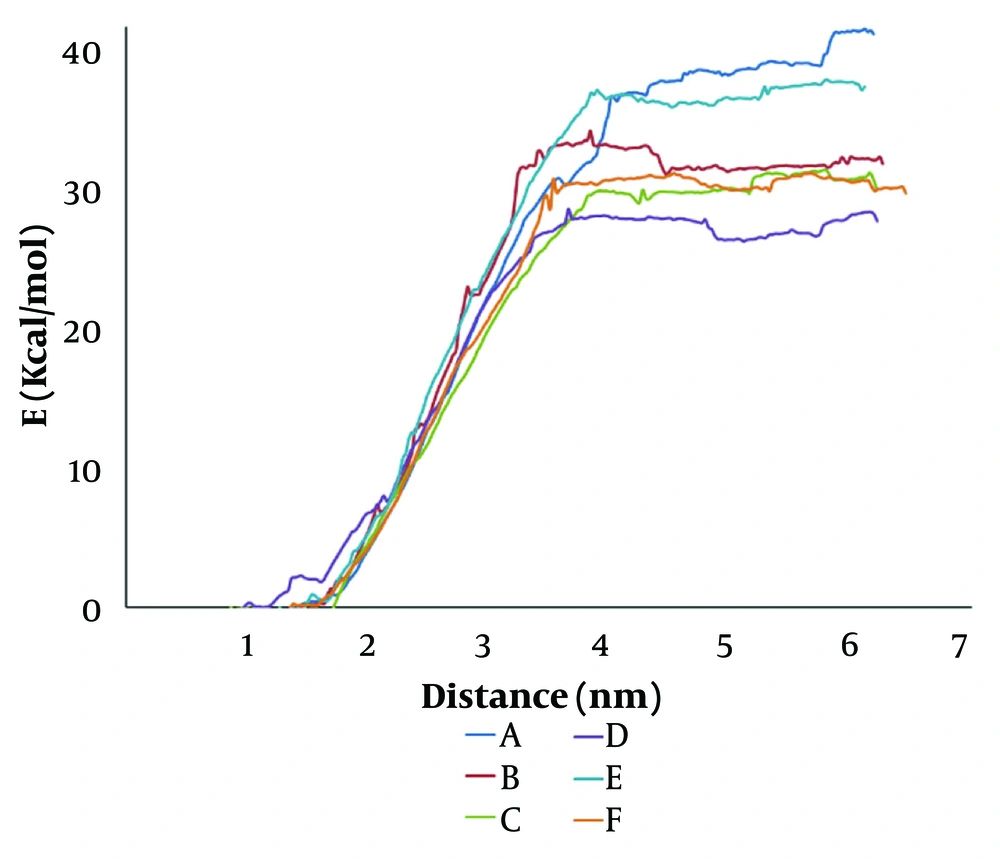
In the PMF/distance curves, the area under the curve represents the total free energy change required to extract a lipid from the bilayer. This cumulative energy provides an alternative means to assess the binding energy (E) reported in Table 1. A larger AUC corresponds to higher binding energy, reflecting a more substantial free energy barrier. Thus, the integrated AUC corroborates the trends observed in the binding energy calculations and offers additional insight into how the energy is distributed across the detachment process. Similarly, in force/time diagrams, the area under the curve represents the total work performed during the lipid extraction process. Since work is defined as the time integral of force, a larger area indicates that more energy is required to overcome the attractive interactions in the membrane. In our dynamic analyses (Figure 1), systems that exhibit higher and more sustained force responses during detachment (such as S1) will have a greater AUC, corresponding well with their higher binding energies and more extended detachment distances.
Potential of mean force (PMF) profiles of lipid detachment in liposomal systems: A, pure 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE); B, 70% DSPE + 30% cholesterol; C, 50% DSPE + 50% 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC); D, 35% DSPE + 35% DSPC + 30% cholesterol; E, 50% DSPE + 50% 1,2-distearoyl-sn-glycero-3-phospho-L-serine (DSPS); F, 35% DSPE + 35% 1,2-distearoyl-sn-glycero-3-phospho-L-serine (DSPS) + 30% cholesterol.
4.2. Comparative Free Energy Landscapes
Figure 2 presents the PMF profiles for lipid detachment as a function of the extraction distance. These profiles graphically underscore the influence of both cholesterol and lipid purity on the free energy landscape of lipid detachment. The significant reduction in barrier height and detachment distance in cholesterol-containing systems indicates a loss in membrane stability.
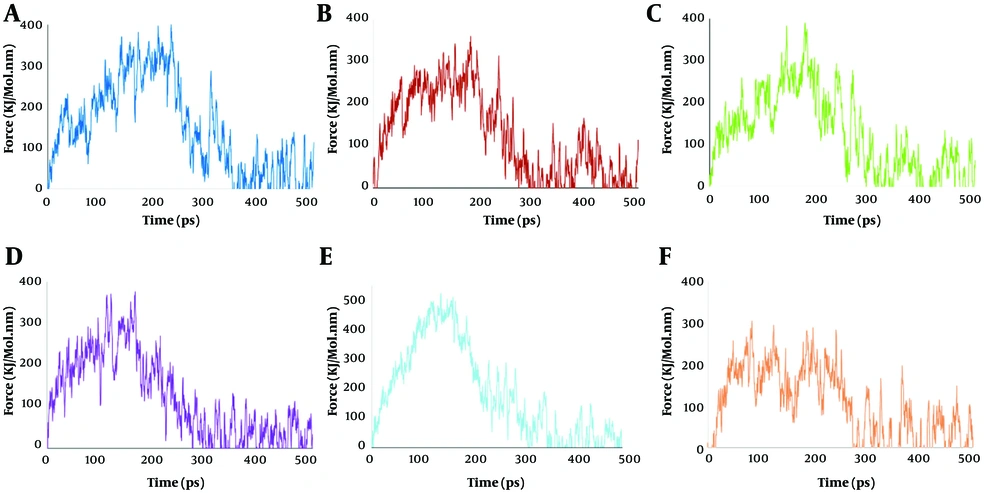
Force VS time diagrams across different lipid compositions: A, pure 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE); B, 70% DSPE + 30% cholesterol; C, 50% DSPE + 50% 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC); D, 35% DSPE + 35% DSPC + 30% cholesterol; E, 50% DSPE + 50% 1,2-distearoyl-sn-glycero-3-phospho-L-serine (DSPS); F, 35% DSPE + 35% DSPS + 30% cholesterol.
4.3. Force versus Time
Figure 3 depicts the force versus time profiles for each of the six systems, capturing the change in the applied forces during lipid extraction. In system S1, the force required to initiate and sustain lipid detachment peaks at a high value and then remains elevated over an extended duration, correlating with the high binding energy and AUC observed in Table 1. This prolonged duration in force application indicates strong lipid anchoring and resists detachment even under continuous mechanical stress.
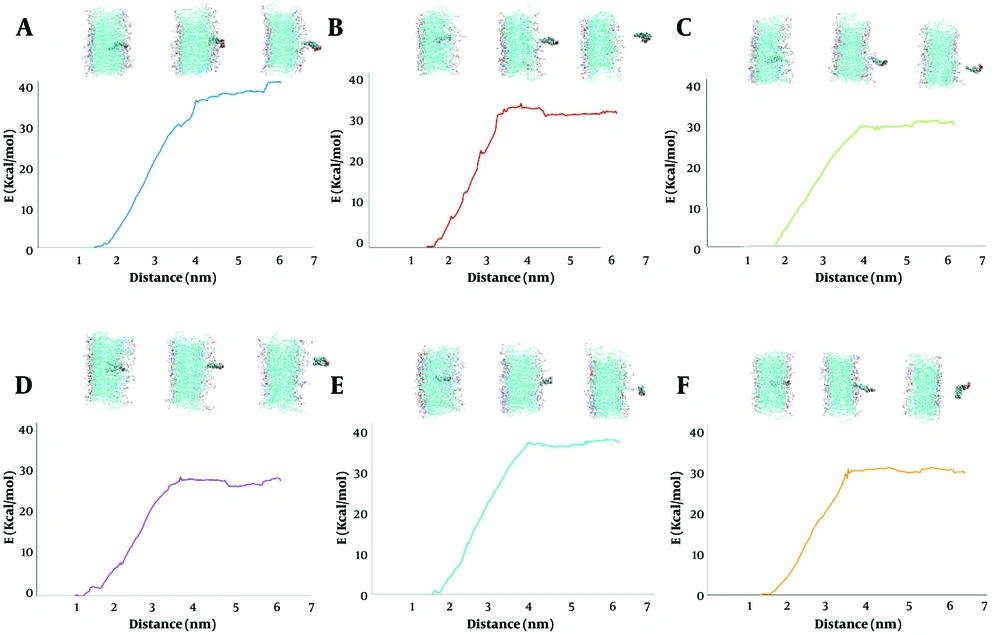
Investigating the conformational changes in the bilayer system, over the PMF profile for different lipid compositions: A, pure 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE); B, 70% DSPE + 30% cholesterol; C, 50% DSPE + 50% 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC); D, 35% DSPE + 35% DSPC + 30% cholesterol; E, 50% DSPE + 50% 1,2-distearoyl-sn-glycero-3-phospho-L-serine (DSPS); F, 35% DSPE + 35% DSPS + 30% cholesterol.
In comparison, the force profile for system S2 demonstrates a lower maximum force and a shorter enforcement phase, consistent with a lower binding energy and reduced AUC. The DSPE/DSPC and DSPE/DSPS systems similarly demonstrate that cholesterol incorporation results in decreased peak forces and shorter periods of elevated force compared to their cholesterol-free counterparts. The lower force requirement for detachment implies that lipid anchors (and any attached targeting ligands) may be more susceptible to dislodgement under shear stress, which is undesirable for targeted drug delivery systems.
4.4. Correlation of Membrane Conformation with Energetic Profiles
Figure 1 integrates PMF curves with representative micrographs or schematic depictions of the corresponding membrane conformation schematics. In system S1, the images reveal a highly ordered, densely packed bilayer where DSPE molecules are uniformly arranged and deeply embedded. This structural order is aligns with the high binding energy in the PMF profile.
Conversely, the images for system S2 show a noticeably disordered membrane with cholesterol-induced microdomains. In these regions, the lipid packing is disrupted, and lipids are more shallowly embedded. This conformational change is directly correlated with a lower free energy barrier. The figure clearly shows that the deeper and more ordered the lipid embedding within the bilayer, the greater the energy required for detachment. This correlation is essential for predicting the stability of lipid anchors used to tether large targeting molecules.
A particularly intriguing observation arises when comparing the mixed systems: The DSPE/DSPC mixture (S3) exhibits a binding energy considerably lower than that of the DSPE/DSPS mixture under cholesterol-free conditions. This comparison indicates that the presence of DSPC in the bilayer results in weaker overall lipid-lipid interactions compared to DSPS. The DSPC, with its phosphocholine headgroup, contributes to a more ordered but sterically constrained packing environment, whereas DSPS, which has a negatively charged phosphoserine headgroup, supports stronger localized hydrogen bonds and electrostatic interactions that reinforce the bilayer.
However, these interactions may also introduce repulsion, potentially affecting the uniformity of the bilayer. Notably, upon cholesterol incorporation, the decrease in binding energy is more pronounced in the DSPS-containing system. For S5, the binding energy decreases from 38.19 kcal/mol to 31.43 kcal/mol in S6, a reduction of 17.69%. In contrast, for the DSPE/DSPC system, the binding energy drops from 31.66 kcal/mol in S3 to 28.83 kcal/mol in S4, a reduction of 8.94%. These findings indicate that although the DSPS-based system initially exhibits a higher binding energy than the DSPC-based system, it is also more adversely affected by cholesterol. This greater sensitivity suggests that the stabilizing interactions present in DSPS, such as hydrogen bonding and electrostatic attractions, may be disrupted more dramatically by cholesterol’s insertion, leading to a larger proportional loss of stability.
Another key bilayer organization parameter is the area per lipid. Generally, a lower area per lipid suggests more efficient packing and closer molecular contact between adjacent lipids, which enhances van der Waals and hydrophobic interactions. In the case of DSPS, the smaller effective headgroup, facilitated by its capacity to form hydrogen bonds and stabilize via electrostatic interactions, would be expected to result in a reduced area per lipid relative to DSPC. This structural difference is consistent with the observed lower binding energy in DSPE/DSPC systems, as looser packing typically requires less energy to perturb the bilayer structure and extract a lipid molecule. In summary, the DSPE/DSPS system benefits from the ability of the phosphoserine headgroup to reduce the area per lipid, resulting in a more tightly packed and homogeneous membrane that exhibits higher binding energy compared to the DSPE/DSPC system. This indicates that even though DSPS-containing membranes may be more stable under basal conditions, their strong intermolecular interactions also make them more sensitive to any disruptive effects. Understanding these distinctions between DSPC and DSPS is critical for the rational design of liposomal drug delivery systems. The choice of co-lipid can significantly influence the balance between membrane rigidity and flexibility, which in turn affects both the encapsulation efficiency and the retention of surface-bound targeting ligands. When the goal is to maximize the stability of the targeting interface, as is often required for prolonged circulation and enhanced therapeutic efficacy, choosing a co-lipid that promotes tighter packing (i.e., DSPS) appears beneficial.
4.5. Conclusions
This study employed all-atom MD simulations combined with umbrella sampling to unravel the molecular determinants of lipid detachment from liposomal membranes. Our investigation focused on six liposomal systems with varying phospholipid compositions and cholesterol content, aiming to optimize the stability of these membranes for targeted drug delivery applications. The pure DSPE system displayed the highest binding energy, indicative of strong intermolecular interactions stemming from a homogeneously packed, saturated lipid environment. In contrast, the addition of 30% cholesterol led to a reduction in binding energy in all systems. These results underline cholesterol’s disruptive effect on lipid packing, resulting in a lower energy barrier. The DSPE/DSPC mixture exhibited a binding energy of 31.66 kcal/mol, which is considerably lower than the 38.19 kcal/mol observed in the DSPE/DSPS system. In DSPE/DSPC membranes, binding energy decreases from 31.66 to 28.83 kcal/mol (an 8.94% reduction), whereas in DSPE/DSPS membranes, it drops more significantly from 38.19 to 31.43 kcal/mol (a 17.69% reduction). These findings provide crucial insights into how lipid composition and cholesterol content can be fine-tuned to design liposomes with optimized stability and performance for drug delivery. By understanding the molecular interactions driving lipid detachment, it is possible to design liposomal formulations that balance membrane rigidity with the flexibility required for controlled drug release and efficient cellular targeting. Ultimately, our study establishes a comprehensive framework for optimizing liposomal formulations by carefully balancing lipid purity and cholesterol content. Future work should incorporate experimental validation and consider additional physiological parameters to further refine the design of targeted nanocarriers, thereby enhancing therapeutic efficacy in clinical applications.